Photoactivated Localization Microscopy
Principle, Procedures and More
Overview
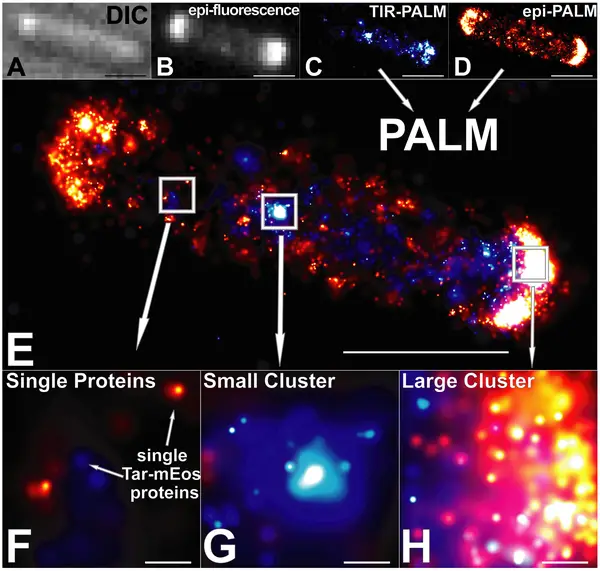
Photoactivated localization microscopy (PALM), developed by Eric Betzig and Harald Hess in the mid-2000s, is a form of super-resolution fluorescence microscopy that allows for highly resolved imaging beyond the diffraction limit of typical optical microscopes. As such, it allows for the visualization of single molecules at about 10 times the resolution of ordinary microscopes.
While ordinary microscopes (compound microscopes used in many laboratories etc) are useful for studying various dynamic events and cellular structures etc, the diffraction limit makes it difficult to study biological processes at the molecular scale.
As compared to these microscopes, Photoactivated localization microscopy allows for 10-50 nanometer resolution which allows for the resolution of spatial details of tightly packed molecules. Through this technique, then, it's possible to study and describe such concepts as molecular interactions etc.
* Super-resolution microscopy techniques include a number of microscopy methods to overcome/break the diffraction limit by temporally/spatially modulating the excitation light.
These techniques are divided into two main groups, these include:
· Deterministic super-resolution - E.g. Stimulated emission depletion (STED) microscopy and Ground-state depletion microscopy.
· Stochastic super-resolution - E.g. Vertico spatially modulated illumination, Super-resolution optical fluctuation imaging (SOFI), and Photoactivated localization microscopy.
Given that cellular structures in the body are made up of biomolecules at the nanometer scale (e.g. tubulins that make up microtubules measure about 8nm), nanometer resolution is required to get clear visualization and thus an opportunity to study these structures.
Today, this is achieved with the use of super-resolution microscopy techniques. This becomes problematic when using a light microscope given that in order to resolve two points as distinct from each other, they have to be more than 200nm apart from each other.
Diffraction Limit
In order to understand how Photoactivated localization microscopy achieves higher resolution beyond the diffraction limit, it's important to understand what the diffraction limit is and how it affects ordinary optical microscopes.
In his studies during the 1870s, Ernst Abbe, a German physicist, noticed that the resolution of an optical microscope is dependent on the aperture of its optics as well as the wavelength of light.
Today, it's well understood that for the traditional forms of fluorescence microscopy, diffraction of light passing through the lenses and the circular apertures of the optics results in resolution limitations.
Because of the wave-like characteristics of diffracted light, objects that are smaller than about 200 nanometers in lateral dimensions and 500 nanometers in axial dimensions cannot be clearly visualized because they end up as blurs.
As a result, it's difficult to clearly visualize the majority of sub-cellular structures due to the fact that they are too small in size (smaller than the dimensions described).
* The resolution limit of ordinary light microscopes is also known as point-spread function (PSF).
* As a result of diffraction (scattering of light), a microscope's ability to distinguish between objects divided by a lateral distance that is less than the wavelength of light being used to image is limited.
This means that in a scenario where the distance between two objects is less than half the wavelength of the light being used, diffraction negatively affects the ability of a microscope to distinguish between the objects. For this reason, an individual cannot clearly visualize the two distinct objects.
Principle
Essentially, Photoactivated localization microscopy achieves its purpose through the imaging of single fluorophores as well as regulated activation and sampling of sparse subsets of the labels sequentially. Here, it's worth noting that fluorophores have a number of properties that can be exploited for photoactivated localization microscopy.
For instance, in addition to being photoswitchable, they are also reversible and photoactivatable.
When exposed to given wavelengths, a change in emission spectra is observed (e.g. becoming fluorescent and changing from one state of fluorescence to another). In practice, the basic principle behind the technique is that an activating laser beam (low-power) stochastically converts the fluorophores. As a result, just a few fluorophores are in the active state at a given time.
Following the initial activation, they are then imaged using an illuminating laser beam (high-power) which rapidly reverts them to the inactive state. To ensure that all molecules are imaged, this process is repeated several thousand times in order to reconstruct the image.
Procedure
The following is a brief procedure for Photoactivation localization microscopy of proteins:
To visualize the sample at the nanoscopic level, the sample is first labeled with photo-activatable (or switchable) fluorescent molecules (fluorophores). Here, some of the molecules used for this purpose include PA-GFP (photoactivatable green fluorescent protein), PA-TagRFP (photoactivatable red fluorescent protein), Dreiklang, Dronpa, and PA-mCherry1 among others.
Once the sample (protein) is labeled, it's then illuminated using a given wavelength of light which causes the probes (photoactivable) to go through an on/off (active/inactive) state.
To overcome the issues of diffraction, the fluorescent molecules are distinguished from each other through activation state or distinct emission spectra. For instance, through illumination, random, sparsely distributed fluorescent proteins are photoactivated at any given time to produce an image of the activated molecules.
These molecules are separated by a distance that is greater than the PSF (point-spread function) of the microscope that would otherwise cause blurring due to diffraction.
* Typically, two point sources with PSFs significantly overlapping at the image place cannot be resolved using conventional microscopes.
Here, then, one of the biggest advantages of Photoactivated localization microscopy is the fact that random (stochastically) and sparse fluorescent proteins are photoactivated at any given time.
In doing so, molecules separated by a distance that is greater than the PSF are photoactivated and imaged while avoiding the problem of diffraction limit.
A low-power activating laser beam is first used to convert the sparse fluorophores into an active state while a higher-power illuminating laser beam serves to image them. This causes the fluorophores to revert back to the inactive state before the process can be repeated.
Inactivating the imaged molecules (through photobleaching) ensures that the molecules (which have already been localized/imaged) are removed from the set of inactivated molecules. This is particularly important as it ensures that they are not imaged again with consequent imaging of the other molecules (inactivated molecules) which in turn allows for proper reconstruction of the whole image (by imaging random molecules at a time). This also helps prevent diffraction.
Once the subset of molecules is photobleached, a new sparse subset is activated (using the low-power activating beam) and imaged (which again causes photobleaching of the new subset of molecules).
By repeating the process over thousands of frames, all the molecules in the field of view are imaged and compiled so as to reconstruct or build up a super-resolution image. Given that the technique operates by localizing single molecules, it's possible for researchers to study given processes with high precision.
Some of the main applications of PALM today include:
· HIV studies - Using Photoactivated localization microscopy, it has become easier for researchers to identify and categorize the Gag polyprotein responsible for the assembly process of HIV-1 and also differentiate HIV-1 from HIV-2
· Study the architecture of various organelles - While it's possible to identify different cell organelles using ordinary microscopes, using PALM has allowed researchers to get a better understanding of the structural architecture of such organelles as mitochondria.
For instance, using PALM microscopy, researchers were able to closely study the cytochrome C oxidase system in mitochondria thus proving a better understanding of the spatial arrangement in the structure.
· Focal adhesion - Because of the high resolution that PALM provides, it has become possible to study and get a better understanding of interaction/adhesive contact between cells and the extracellular matrix. This is particularly important in that it allows researchers to study and describe any pathological process affecting such interactions.
Interferometric Photoactivated Localization Microscopy (PALM)
The Interferometric photoactivated localization microscopy is a technique that combines the strengths of photoactivated localization microscopy (PALM) and single-photon, simultaneous multiphase interferometry making it possible to visualize the three-dimensional structure of molecules.
For researchers, this technique has a number of benefits that include:
- Allows for the visualization of three-dimensional structures of molecules
- Molecules are visualized at a nanometer-length scale (makes it possible to get a very close look at the molecules)
- Provides high accuracy of proteins at the nanoscopic scale (nanometer scale)
- Axial resolution of this technique is higher than that of other super-resolution techniques
By combining the single-molecule localization aspect of PALM and multi-phase interferometry, it has become possible to achieve a nearly isotropic resolution in both the lateral and axial directions. For this reason, the technique is typically used for studies that require microscope resolution near the electron level.
* While this technique has been praised for the high spatial resolution it provides, the temporal resolution offered is more limited when compared to the other super-resolution techniques. For this reason, fixed samples are often used.
How iPALM works
In order to achieve high resolution using iPALM, this approach is dependent on the features of single-molecule localization (of PALM) as described above.
In order to achieve axial resolution, the technique has to make up interferometry principles where a light beam is split into several paths (two or three) before being recombined so as to form interference signals. Using features of the signal, it's possible to compute the difference in path length to within nanometers.
For the paths for fluorescent signal to traverse, this technique employs the use of two objectives for sample imaging (where one of the objectives is at the top while the other is located at the bottom). This allows for most (if not all) the emitted fluorescence from the sample to be collected this contributing to the photo-efficiency of the technique.
The light below and above the sample is then collected by the PALM microscope feature (adapted in a manner that allows it to collect the light) and travels to towards a beam splitter which in turn sends the beam (recombined) to a set of three different cameras.
The amount of light reaching the camera is largely dependent on the depth of the molecule within the sample. Using the cameras, the image is recorded making it possible to determine the height of the molecule.
Fluorescence Photoactivation Localization Microscopy (FPALM)
Like Photoactivation localization microscopy and Stochastic optical reconstruction microscopy, Fluorescence photoactivation localization microscopy is also an individual molecule localization technique used to visualize samples (both biological and non-biological) with subdiffraction-limited resolution by using lasers to excite photo-switchable protein fluorophores on the surface of the sample.
While fPALM uses the same imaging procedure as that used in Photoactivation localization microscopy, they are different in that Fluorescence Photoactivation localization microscopy uses aTotal internal reflection fluorescence microscope while Photoactivation localization microscopy typically uses a confocal microscope.
For the fPALM, however, a widefield fluorescence objective with his NA (numerical aperture) can be used because it does not limit the focal place to the proximity of a coverslip. Through this approach, it's possible to image a three-dimensional sample within a single focal place that has a thickness of equal depth of field. In the process, lateral resolution of tens of nanometers within a plane is achievable.
* As is the case with PALM, the image of molecules distributed in living and fixed cells are captured through a cycle of activation, localization, and photo-bleaching.
Two of the most beneficial properties of this technique include:
· Allow for high spatial resolution - High spatial resolution ensures that the images are clear which makes it easier to study the sample
· High speed of image acquisition - This means that the rate of molecule localization achieved using fPALM is high which makes it possible to obtain a complete image of the sample within a short period of time. One of the biggest benefits of this is that it significantly reduces the impact of sample drift which in turn allows for imaging of live cells
Cryo-PALM
While PALM imaging of single molecules was first demonstrated at cryogenic temperatures (temperatures below -150 degrees Celsius), It's often carried out at room temperatures today. This is largely because of the absence of fluorescent proteins that are phototransformable at such low temperatures.
However, using cryo-PALM-CET, a relatively new technique, it's possible to image frozen specimen. Here, cells are first preserved by vitrification thus limiting cryo-PALM probes to ones that can be incorporated into live cells. In most cases, the fluorescent proteins are negatively affected by the low temperatures with the exception of a few (E.g. PA-GFP that can be used).
One of the other issues that can affect this technique is that exposure to intense laser can warm the specimen (frozen specimen) resulting in the crystallization and denaturation of the sample. To overcome this problem, the sample can be exposed to a laser with just enough energy to activate the fluorophore and image the specimen.
See also: Cryo-Electron Microscopy and Cryo-Electron Tomography
Return to Fluorescence Microscopy
Return to Super-Resolution Microscopy
Return to Widefield Epifluorescence Microscopy
Return to Microscopy Imaging Techniques main page
Return from Photoactivated localization microscopy to MicroscopeMaster home
References
Catherine G. Galbraith and James A. Galbraith. (2011). Super-resolution microscopy at a glance. Cell Science at a Glance.
Haining Zhong. (2010). Photoactivated Localization Microscopy (PALM): An Optical Technique for Achieving 10-nm Resolution. ResearchGate.
Prabuddha Sengupta and Jennifer Lippincott-Schwartz. (2012). Quantitative Analysis of Photoactivated Localization Microscopy (PALM) Datasets Using Pair-correlation Analysis. ncbi.
Samuel T. Hess et al. (2018). Ultra-High Resolution Imaging of Biomolecules by Fluorescence Photoactivation Localization Microscopy (FPALM). ncbi.
Yi-Wei Chang et al. (2014). Correlated cryogenic photoactivated localization microscopy and electron cryotomography. ResearchGate.
Links
https://oni.bio/palm-microscopy
https://insights.oni.bio/blog/the-best-fluorophores-for-palm-imaging
Find out how to advertise on MicroscopeMaster!
![ELYRA-superresolution structured illumination, photoactivated localization microscopy,both technologies simultaneously-ZEISS Microscopy-Germany[CC BYSA(https://creativecommons.org/licenses/by-sa/2.0)] ELYRA-superresolution structured illumination, photoactivated localization microscopy,both technologies simultaneously-ZEISS Microscopy-Germany[CC BYSA(https://creativecommons.org/licenses/by-sa/2.0)]](https://www.microscopemaster.com/images/zeisssuperresolutionmicroscope.jpg)